Genetic information is encoded in the deoxyribonucleic acid (DNA). In form of a long double-helix molecule, lo-cated in living cells, it governs most of the organisms traits. Explicitly, information from genes is used to form func-tional gene products such as proteins. This process of gene expression is used by all known forms of life on earth to generate the macromolecular machinery for life. Thus, it poses the fundamental level of how the genotype causes the phenotype, i.e. the composite of organisms’ observ-able characteristics. Genomic modification is a powerful tool to amend those characteristics. Reproductional and environmentally caused changes to the DNA is a substrate for evolution. In nature, those changes happen and may cause favourable or unfavourable changes to the phenotype, which allow the cell or organism to improve or reduce the ability to survive and reproduce, respectively.
In the first half of the 20th century, several methods to alter the genetic structure of cells were discovered, which include exposing it to heat, X-rays, UV-light, and chemicals1-4. A significant number of crop cultivated today were developed using those methods of traditional muta-genesis, an example of which is Durum wheat, the most prevalent wheat for pasta production. With traditional mu-tagenesis thousands of mutations are introduced at random within the DNA of the plant. A subsequent screening iden-tifies and separates cells with favourable mutations in their DNA, followed by attempts to remove or reduce possible unfavourable mutations in those by mutagenesis or cross-breeding.
As those methods are usually unspecific and complex, researchers have developed site-determined gene editing techniques, the most successful of which is the so called CRISPR/Cas9 method (clustered regularly interspaced short palindromic repeats). This method borrows from how bacteria defend viral invasion.6 When the bacterium detects virus DNA invasion, it forms two strands of RNA (single helix molecules), one of which contains a sequence that matches that of the invading virus DNA and is hence called guide RNA. These two RNAs form a complex with a Cas9 protein, which, as a nuclease enzyme, can cleave DNA. When the guide RNA finds the target in the viral genome, the RNA-Cas9 complex will lock to a short se-quence known as the PAM, the Cas9 unzippes the viral DNA to which the RNA will match. Cas9 then cleaves the viral DNA, forcing the cell to repair the DNA.6 As this repair process is error prone, it may lead to mutations that might disable certain genes, changing the phenotype. In 2012 and 2013 it was discovered that the guide RNA can be considerably modified for the system to work site-determined5, and that by modifying the enzyme it not only works in bacteria and archaea, but also in eukaryotes (plants and animals), respectively.7
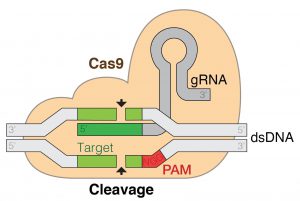
Figure 1: CRISPR/Cas9 working principle.8
Research published since demonstrated the method’s poten-tial for RNA-programmable genome editing. Modifications can be made so during the repair an artificially designed DNA sequence pairs with the cleaved ends, recombines and replaces the original sequence, introducing new genes to the genome.11,12 The advantages of this technique over tra-ditional gene editing methods is multifold. It can act very targeted, i.e. site- and therefore gene-specific in any form of known life. It is comparatively inexpensive, simple enough to be conducted in basic labs, effective, and fast regarding preparation and realisation. The production of multiplex ge-netically modified mice, for instance, was reduced from up to two years to few weeks,9 as CRISPR/Cas9 has the unique advantage over earlier genome editing methods, that multi-plexable targeting is easily achieved by co-expressing Cas9 with multiple single-guide RNAs simultaneously. Conse-quently, within few years after its discovery, it evolved to be the routine procedure for genome modification of virtually all model plants and animals.
The availability of such a method evokes medical and botanical development interests. A plethora of possible medical applications are discussed and researched, among which is healing cancer or treating genetic disorders. For cancer research it is imaginable to induce a multitude of deliberate mutations to artificially form cells similar to can-cerous cell, study the caused modification to the cells, and thus learn to inhibit their reproduction or the original muta-tion. In the clinical research focus now are blood diseases or those related to haematopoietic cells, such as leukaemia, HBV, HIV, or haemophilia.13,14 This is because for the treatment of those diseases, the cells (blood cells or bone marrow) can be extracted from the body in a known way, their genome can be edited in vitro by the CRISPR/Cas9 method, and finally the cells can be reintroduced to the body. The advantage of the extraction is that no additional vector (agent to help finding the right cells in vivo) is re-quired, and the genomic modification can be controlled ex vivo. While the editing efficiency with CRISPR-Cas9 can be extremely high, the resulting cell population will be inherently heterogeneous, both in the percentage of cells that were edited and in the specific genotype of the edited cells. Potentially problematic for in vivo application is the bacterial origin of the endonuclease Cas9. A large portion of humans show humoral and cell-mediated immune re-sponses to the Cas9 protein complex,10 most likely because of prior infection with related bacteria.
Although clinical applications of CRISPR/Cas9 grab a lot of media attention, agricultural applications draw even more commercial interest. Prospects here are the faster, cheaper and more targeted development of crops than by traditional methods of mutagenesis, which are extremely more aggressive in comparison. The main aim is unchanged though: improve plants regarding yield, resistance to dis-eases or vermin, and resilience to aridity, heat, cold, humid-ity, or acidity.15,16 CRISPR/Cas9 is therefore considered an important method to ameliorate agricultural food produc-tion to feed the earth’s ever-growing human population.
Regulations of thusly modified products vary largely be-tween countries. While Canada considers such plants equal to not genetically modified if no transgene was inserted, the USA assesses CRISPR plants on a case by case basis, gauging whether the modification would have been possible by natural mutation. This way they chose to not regulate mushrooms that do not turn brown and maize with an al-tered starch contend. Last year the European court of justice ruled all CRISPR/Cas9 modified plants as genetically mod-ified organisms, reasoning that the risks of such a novel method are unknown, compared to traditional mutagenesis as an established method of plant breeding.
Instigated by genome editing in human-embryonic cells in 201518 a group of scientists called for a moratorium to dis-cuss the possible risks and impact of the wide usage of the CRISPR/Cas9 technology, especially when it comes to mu-tations in humans.19 On the 2015 International Summit on Human Gene Editing leading international scientists con-sidered the scientific and societal implications of genome editing. The discussed issues span clinical, agricultural and environmental applications, with most attention focused on human-germline editing, owing to the potential for this application to eradicate genetic diseases and, ultimately, to alter the course of evolution. Some scientists advise to ban CRISPR/Cas9 based human genomic editing research for the foreseeable future, whereas others favour a rapid progress in developing it.20 A line of argument of support-ers of the latter viewpoint is, that the majority of ethical concerns are effectively based on methodical uncertainties of the CRISPR/Cas9 method at its current status, which can be overcome only with extensive research. Those methodical uncertainties include possible cleavage at undesired sites of the DNA, or insertion of wrong sequences at the cleavage site, resulting in the disabling of the wrong genes or even the creation of new genetic diseases.
Whilst a total ban is considered impractical because of the widespread accessibility and ease of use of this technology,21 the summit statement says, that “It would be irresponsible to proceed with any clinical use of germline editing unless and until (i) the relevant safety and effi-cacy issues have been resolved . . . and (ii) there is broad societal consensus about the appropriateness of the pro-posed application.” The moral concerns about embryonic or germline treatment base on the fact that CRISPR/Cas9 not only would allow the elimination of genetic diseases, but also enable genetic human enhancement, from simple tweaks like eye colour or non-balding to severe modifica-tions relating bone density, muscular strength or sensory and mental capabilities.
Although most scientist echo the summit statement, in 2018 a biochemist claimed to have created the first genetically edited human babies, two twin sisters. After in vitro fertil-ization, he targeted a gene that codes for a protein that one HIV variant uses to enter cells, enforcing a kind of HIV immunity, which is a very rare trait among humans.22 His conduct was harshly criticised in the scientific community, widely condemned, and-after enormous public pressure-redoing forbidden by the responsible regulatory offices.
Ultimately the CRIPSR/Cas9 technology is a paramount example of real world societal implications of basic re-search and demonstrates researchers’ responsibilities. This also raises the question whether basic ethical schooling should be part of every researcher’s education.
— Alexander Kronenberg
Read more:
[1] K. M. Gleason (2017) “Hermann Joseph Muller’s Study of X-rays as a Mutagen”
[2] Muller, H. J. (1927). Science. 66 (1699): 84–87.
[3] Stadler, L. J.; G. F. Sprague (1936). Proc. Natl. Acad. Sci. U.S.A. US Department of Agriculture and Missouri Agricul-tural Experiment Station. 22 (10): 572–8.
[4] Auerbach, C.; Robson, J.M.; Carr, J.G. (March 1947). Sci-ence. 105 (2723): 243–7.
[5] M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna, E. Charpentier. Science, 337, 2012, p. 816–821.
[6] R. Sorek, V. Kunin, P. Hugenholtz. Nature reviews. Micro-biology. 6, 3, (2008), p. 181–186.
[7] Cong, L., et al., (2013). Science. 339 (6121) p. 819–823.
[8] https://commons.wikimedia.org/wiki/File:GRNA-Cas9.png
[9] H. Wang, et al., Cell. Band 153, 4, (2013), S. 910–918.
[10] D. L. Wagner, et al., Nature medicine. (2018).
[11] O. Shalem, N. E. Sanjana, F. Zhang; Nature reviews. Genet-ics 16, 5, (2015), p. 299–311.
[12] T. R. Sampson, D. S. Weiss; BioEssays 36, 1, (2014), p. 34–38.
[13] G. Lin, K. Zhang, J. Li; International journal of molecular sciences 16, 11, (2015), p. 26077–26086.